Abstract
The myelodysplastic/myeloproliferative neoplasms (MDS/MPN) are a unique group of hematologic malignancies characterized by overlapping features of myelodysplastic syndromes and myeloproliferative neoplasms. The category includes atypical chronic myeloid leukemia (aCML), chronic myelomonocytic leukemia (CMML), MDS/MPN, unclassifiable (MDS/MPN, U), and MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T). The recent adaptation of next generation sequencing in genetic testing increased the knowledge of the molecular pattern of MDS/MPN. However, no specific gene signatures have been identified and underlying transcriptional mechanisms are still poorly understood. Therefore, we analyzed the transcriptome of a cohort of 49 aCML, 102 CMML, 50 MDS/MPN-U, and 72 MDS/MPN-RS-T patients, morphologically defined according to the WHO classification.
Total RNA was used for 150bp paired-end RNA sequencing with a median read depth of 50 million. The edgeR package was used to perform a combined normalization and differential expression (DE) analysis of the obtained estimated gene counts by integration of trimmed mean of M-values normalization factors into the statistical model used to test for DE. Genes with an FDR < 0.05 and an absolute logFC > 1.5 were considered DE. 273 unique genes were identified differentially expressed between the entities. We found that the transcriptional profile of CMML patients was dominated by an upregulation of MAFB compared to all other entities (p < 0.001). MAFB encodes a transcription factor which is specifically expressed in myeloid cells and a master regulator of human monocytopoiesis.
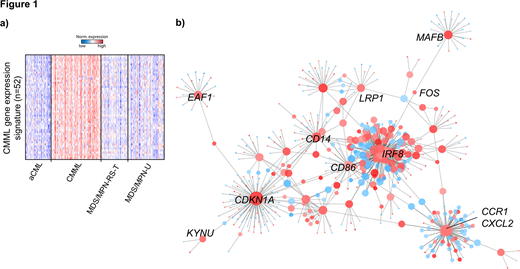
In order to unravel the potential effects of MAFB regulation we performed Spearman's correlation analysis to identify co-expressed genes. 51 genes showed a similar expression (correlation coefficient > 0.7) as MAFB, establishing a CMML gene expression signature (GES) (Figure 1a). The GES included the cell cycle inhibitor CDKN1A, the transcription factors IRF8, EGR2, and MYCL, and the cell surface receptor LRP1. LRP1 represents a therapeutic target and has recently been identified as a key factor in a network for multi-cancer clinical outcome prediction. MAFB was also co-expressed with KYNU, which is synthesized by indoleamine 2,3-dioxygenase, an enzyme expressed by leukemia blasts, and which has been linked with unfavorable outcome in hematologic malignancies. MAFB and KYNU were also co-expressed with multiple CD markers, which are often used as targets of immunotherapy approaches in cancer. High expressions of KYNU can block the anti-tumor effects of chimeric antigen receptor T-cell therapy, indicating complex interactions between the co-expressed genes.
To better understand these underlying molecular interactions on a more global scale and to expose potential targets for intervention, a transcriptional network of the 51 MAFB co-expressed genes was reverse engineered. We used STRING, a publicly available database of protein-protein interactions, to build the scaffold of the network and subsequently pruned the network based on our gene expression data. The reconstructed, MAFB associated regulatory network (Figure 1b) identified c-FOS as an additional regulatory link. c-FOS is a known oncogene that heterodimerizes with c-JUN, ATF3, and JDP2 to form the structure of the AP-1 complex. The AP-1 complex is a key regulator of multiple biological processes such as cell proliferation and apoptosis. We observed a considerably higher expression of c-FOS compared to c-JUN, ATF3, and JDP2 in CMMLs. The MafB/cFos heterodimer is known to explicitly repress apoptosis.
The co-expression network was significantly enriched for transcriptional activation of p53 responsive genes (p < 0.001). Interestingly, we also found that the p53-regulated long intergenic non-coding RNA lncPRESS1 was down-regulated in our cohort of CMML patients (p < 0.001).
Conclusions: 1) MAFB represents a promising gene expression marker to distinguish CMML pts from other MDS/MPN overlap entities. 2) The identified core regulatory signature in CMML pts showed complex interactions to modulate diverse biological processes and might serve as therapeutic targets.
Walter:MLL Munich Leukemia Laboratory: Employment. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Meggendorfer:MLL Munich Leukemia Laboratory: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal